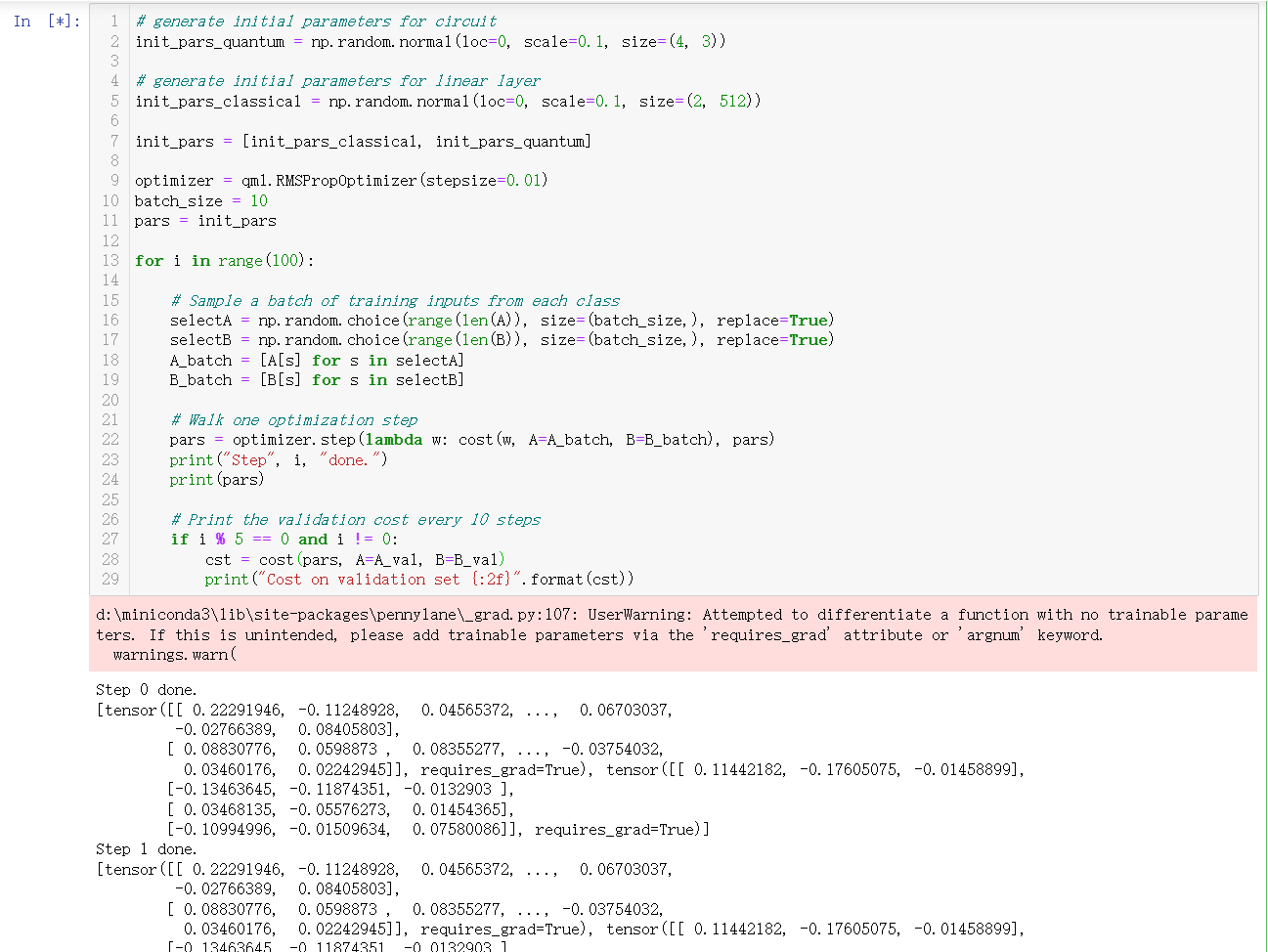
We study the case AA and find that the training is invalid, the trained parameters are the same as the initialized parameters, I wonder why?
Step 0 done.
[tensor([[ 0.22291946, -0.11248928, 0.04565372, …, 0.06703037,
-0.02766389, 0.08405803],
[ 0.08830776, 0.0598873 , 0.08355277, …, -0.03754032,
0.03460176, 0.02242945]], requires_grad=True), tensor([[ 0.11442182, -0.17605075, -0.01458899],
[-0.13463645, -0.11874351, -0.0132903 ],
[ 0.03468135, -0.05576273, 0.01454365],
[-0.10994996, -0.01509634, 0.07580086]], requires_grad=True)]
Step 1 done.
[tensor([[ 0.22291946, -0.11248928, 0.04565372, …, 0.06703037,
-0.02766389, 0.08405803],
[ 0.08830776, 0.0598873 , 0.08355277, …, -0.03754032,
0.03460176, 0.02242945]], requires_grad=True), tensor([[ 0.11442182, -0.17605075, -0.01458899],
[-0.13463645, -0.11874351, -0.0132903 ],
[ 0.03468135, -0.05576273, 0.01454365],
[-0.10994996, -0.01509634, 0.07580086]], requires_grad=True)]
Step 2 done.
[tensor([[ 0.22291946, -0.11248928, 0.04565372, …, 0.06703037,
-0.02766389, 0.08405803],
[ 0.08830776, 0.0598873 , 0.08355277, …, -0.03754032,
0.03460176, 0.02242945]], requires_grad=True), tensor([[ 0.11442182, -0.17605075, -0.01458899],
[-0.13463645, -0.11874351, -0.0132903 ],
[ 0.03468135, -0.05576273, 0.01454365],
[-0.10994996, -0.01509634, 0.07580086]], requires_grad=True)]
Step 3 done.
[tensor([[ 0.22291946, -0.11248928, 0.04565372, …, 0.06703037,
-0.02766389, 0.08405803],
[ 0.08830776, 0.0598873 , 0.08355277, …, -0.03754032,
0.03460176, 0.02242945]], requires_grad=True), tensor([[ 0.11442182, -0.17605075, -0.01458899],
[-0.13463645, -0.11874351, -0.0132903 ],
[ 0.03468135, -0.05576273, 0.01454365],
[-0.10994996, -0.01509634, 0.07580086]], requires_grad=True)]
Step 4 done.
[tensor([[ 0.22291946, -0.11248928, 0.04565372, …, 0.06703037,
-0.02766389, 0.08405803],
[ 0.08830776, 0.0598873 , 0.08355277, …, -0.03754032,
0.03460176, 0.02242945]], requires_grad=True), tensor([[ 0.11442182, -0.17605075, -0.01458899],
[-0.13463645, -0.11874351, -0.0132903 ],
[ 0.03468135, -0.05576273, 0.01454365],
[-0.10994996, -0.01509634, 0.07580086]], requires_grad=True)]
Step 5 done.
[tensor([[ 0.22291946, -0.11248928, 0.04565372, …, 0.06703037,
-0.02766389, 0.08405803],
[ 0.08830776, 0.0598873 , 0.08355277, …, -0.03754032,
0.03460176, 0.02242945]], requires_grad=True), tensor([[ 0.11442182, -0.17605075, -0.01458899],
[-0.13463645, -0.11874351, -0.0132903 ],
[ 0.03468135, -0.05576273, 0.01454365],
[-0.10994996, -0.01509634, 0.07580086]], requires_grad=True)]
We found out that the training is ineffective, the parameters generated by each iteration do not change and therefore cannot be dichotomized.
@Maria_Schuld
@manu_manohar
@Andre_Tavares